E.2 Data Management
Version 3.0 February 2021
Download as pdf
E.2.1 Data Management Overview
Data coordination and management have been key strengths of the ICGC to date. ICGC-ARGO is considerably larger in scale than ICGC and involves a far richer and more complex set of clinical and environmental information, which requires structural changes to the existing Data model to ensure the sound management of ICGC-ARGO data through several operational entities described in this policy. The overarching principles of the data management system and its design are:
- Provide secure and reliable mechanisms for the sequencing centres, clinical data managers, and other ICGC participants to upload their data;
- Track data sets as they are uploaded and processed, to perform basic integrity checks on those sets;
- Allow regular audit of the project in order to provide high-level snapshots of the consortium's status;
- Perform more sophisticated quality control checks of the data itself, such as checks that the expected sequencing coverage was achieved, or that when a somatic mutation is reported in a tumour, the sequence at the reported position differs in the matched normal tissue;
- Enable the distribution of the data to the long-lived public repositories of genome-scale data, including sequence trace repositories and microarray repositories;
- Provide essential meta-data to each public repository that will allow the data to be findable and usable;
- Facilitate the integration of the data with other public resources, by using widely-accepted ontologies, file formats and data models;
- allow researchers to compute across data from ICGC ARGO donors that are stored in multiple localities and to return analytic results that span the entire distributed data set.
- support for hypothesis-driven research: The system should support small-scale queries that involve a single gene at a time, a short list of genes, a single specimen, or a short list of specimens. The system must provide researchers with an interactive system for identifying specimens of interest, finding what data sets are available for those specimens, selecting data slices across those specimens
Each data producer will manage its own data submission and be responsible for primary QC, data integrity and protection of confidential information.
E.2.2 ICGC Data Management Infrastructure
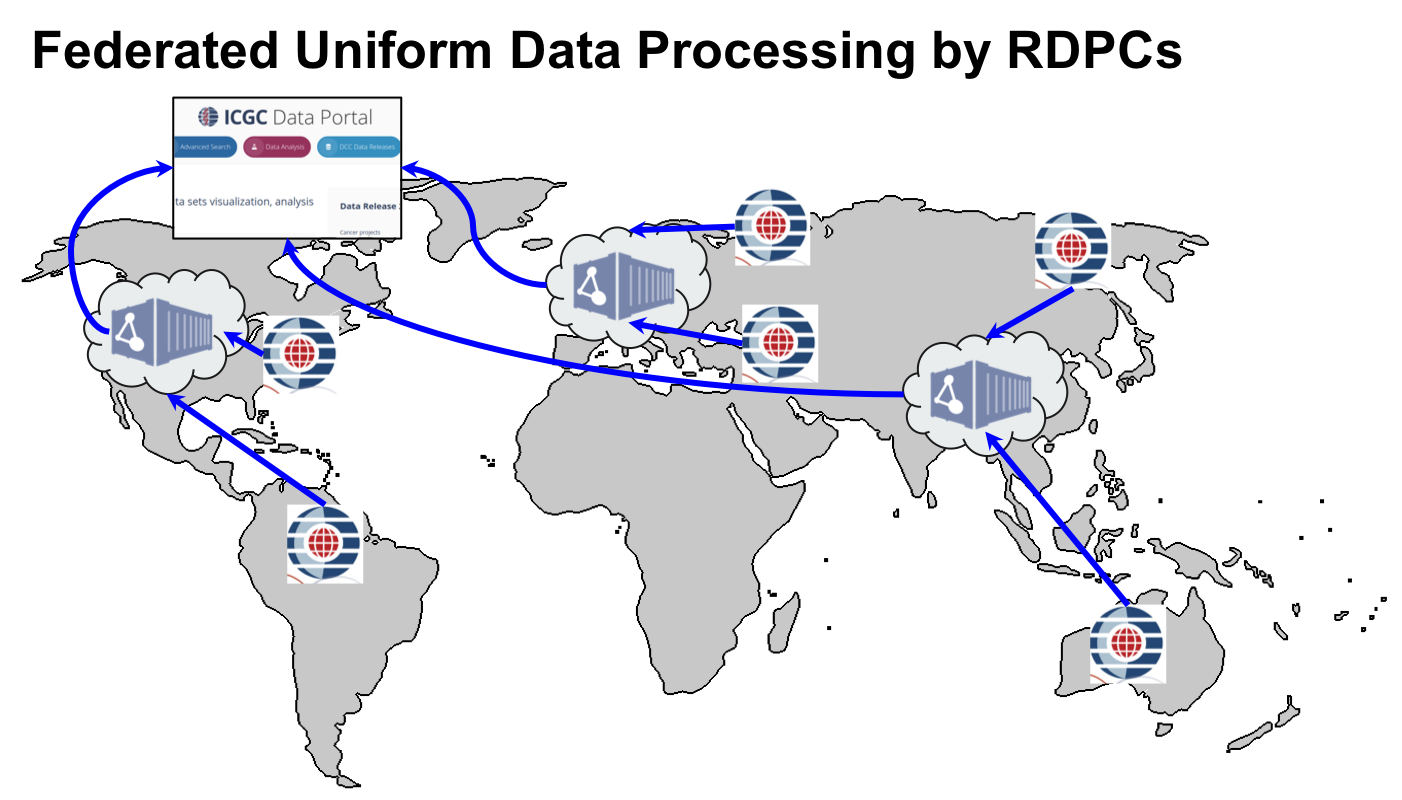
The ICGC-ARGO Data Coordination Centre (DCC) in collaboration with the working groups and consortium members will define the clinical data dictionary and data model. The DCC will provide a submission system for accepting and validating clinical data. The DCC will also coordinate with Regional Data Processing Centre’s (RDPCs) to accept, validate and uniformly analyze molecular data submitted by the ICGC-ARGO sequencing center’s. The RDPCs will process the sequencing data through a standardized series of data analysis pipelines to identify genomic mutations. The use of RDPCs and cloud compute providers together give ICGC-ARGO considerable flexibility in where the data is physically stored. This will allow the project to successfully navigate the changing landscape of international policies on human genetic data storage and distribution. Carefully written software will allow researchers to compute across data from ICGC-ARGO donors that are stored in multiple localities and to return analytic results that span the entire distributed data set.
The interpreted results, along with quality control metrics, will be sent to the DCC for integration with other ICGC-ARGO data sets and dissemination to the scientific and lay communities. Processed sequencing data will be archived in one or more public sequence archives, and mirrored to several cloud compute providers, where qualified researchers will be granted the right to perform additional analyses on the data in a secure and ethically responsible fashion.
The ICGC-ARGO software engineering group will support the operations of the ICGC DCC and the ICGC-ARGO Regional Data Processing Centre’s and will be responsible for developing and distributing the software systems and protocols required for the operation of these Centre’s.
E.2.3 Data Release
POLICY: The members of the International Cancer Genomics Consortium (ICGC) are committed to the principle of rapid data release to the scientific community.
Data producers are recognized to have a responsibility to release data rapidly and to publish initial global analyses in a timely manner. Of equal importance is the responsible use of the data by end-users, which is defined as allowing the data producers the opportunity to publish the initial global analyses of the data within a reasonable period of time, as per the Publication Policy.
The members of the ICGC agree to identify the projects they support and carry out for the comprehensive genomic characterization of human cancers as a set of community resource projects. Data producers, by explicit agreement as members of the ICGC, acknowledge their responsibilities to release data rapidly and to publish initial global analyses in a timely manner. Similarly, funding agencies acknowledge their role in encouraging and facilitating rapid data release from cancer genome projects.
Timing of Data Releases
ICGC ARGO member programs will have privileged access to data from other members of the Consortium based on their level of Membership. Data access is tiered and aimed not to disadvantage Members or Associate Member Data producers, with a framework that encourages data sharing, yet provides data generators with sufficient time to perform analyses:
- Up to 12 months from completion of standardised analyses: Access to Program submitting data only
- 12 months: Access to Full Members
- 18 months: Access to Associate Members
- 24 months: Accessible by external parties
Standardized analyses are considered complete when both mandatory clinical data has been submitted by the Program and molecular data has been uniformly analysed by the RDPCs.